Building and Optimising a custom neural network using evolutionary concepts.
About The Project
As part of an optimisation algorithm module, the task was to develop an algorithm to optimise a problem. I wanted to gain a better understanding of neural networks, more so on how they function. I challenged myself with creating my own neural network class which would be the basis for my optimisation problem. With prior knowledge of implementing genetic algorithms from the first semester, I chose this as the optimiser.
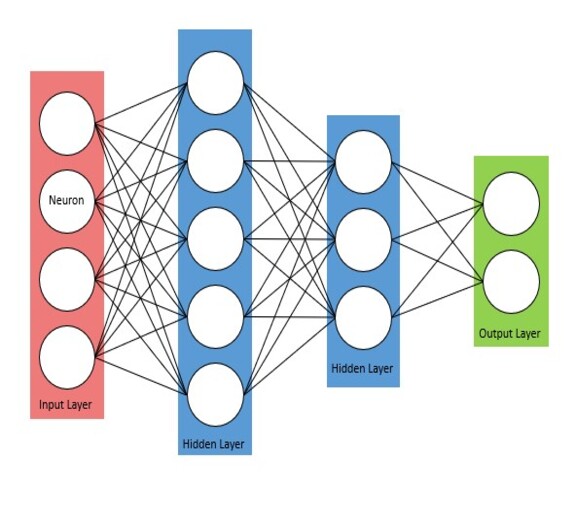
The Neuron and Layer
I approached the problem by reading various literature on how neural networks function. As my knowledge progressed, I implemented what I learnt. This resulted in 2 core components of the network which were a neuron, and layer class.
When initialised, the neuron creates connections of the number of neurons in the next layer. These connections have randomly set weights between 0 and 1 which have a function to obtain them. Each neuron has a weighted sum function which will take an incoming set of weights and inputs, perform a dot product operation and sum the resultant. This value is then passed into a RELU activation function.
The layer is used as a wrapper object for neurons. It has methods to order the weights which will get the first weight of each prior neuron for the first neuron of the current layer and so on. This results in the weights having same dimensions as the inputs from the previous layer, allowing the weighted sum to be performed.
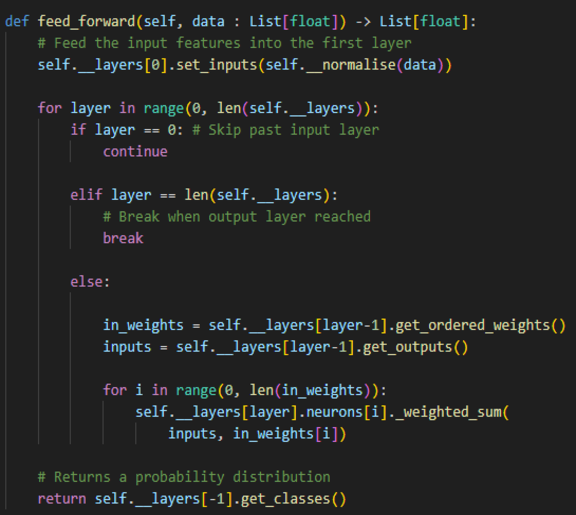
Network Architecture
The neural network classifier class was created such that the number of hidden layers and their sizes can be customised. It automatically scales the input and output layers depending on the number of features given in the training data and number of unique labels.
Once an input has been passed through all layers, the final layer uses a softmax operation to return a set of probabilities normalised between 0 and 1. From this, the loss is calculated using cross entropy between the one hot encoded actual label and the probability vector.
Converting weights to a genetic sequence
The optimiser is based on a genetic algorithm. Each generation of the algorithm will undergo a fitness calculation, selection, crossover and mutation. The heuristic used will be the loss of the network.
To convert the weights into a genome sequence, each weight configuration is unpacked into a one-dimensional array, resulting in each gene representing a single weight. Once the network is ready to be tested or used, the new weights are inserted back into the network in the same way they were unpacked.
Fitness, Selection And Crossover
The fitness for each sequence of weights is measured by summing the loss of all training data in one feed forward and dividing the total loss by 1. This results in high loss sequences having a score tending towards 0 and fitter sequences toward 1.
All sequences are then sorted in a descending order by fitness score. The top 50% of sequences are selected and passed into a crossover function which performs a single-point crossover about the centre-point of the sequence; resulting in two new offspring sequences. This effectively brings the population back to its original size which includes the new sequences and original sequences, maintaining a level of diversity.
Mutation
The mutation function will only mutate a sequence if a threshold has been met. Typically, mutation rates tend to be between 0.01% and 5%, with more frequently occurring mutations the higher the rate. Each gene in the sequence has this chance of mutating. If successful, will add a random value between –2 and 2 to that gene, with a further 2% chance of flipping the sign.
Video Demonstration
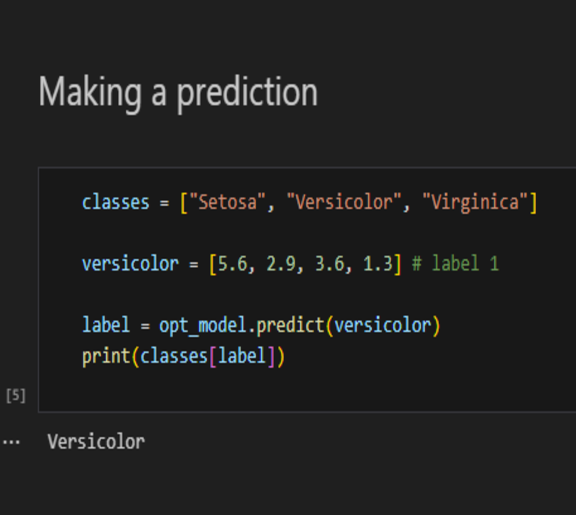
Making Predictions
Once the model has been optimised, the predict method can be used. This will return an integer value which represents the class number. It is important that the same number of features used to train the network is given to the prediction method.
Project Files
An example notebook along with the python files can be found on my github page. Feel free to give it a go!
View Github Repo